Published on 20 February 2022
In a stroke of serendipity, a group of Singapore scientists have identified a new species of bacteria – and named it after Singapore
The staphyloccocus singaporensis (S. singaporensis) bacteria is part of the staphyloccocus aureus (S. aureus) complex. A common bacterial cause of infections, the pathogen may be carried by 20-30% of the population at any given time, said Dr Chew Ka Lip, Consultant, Department of Laboratory Medicine, NUH.
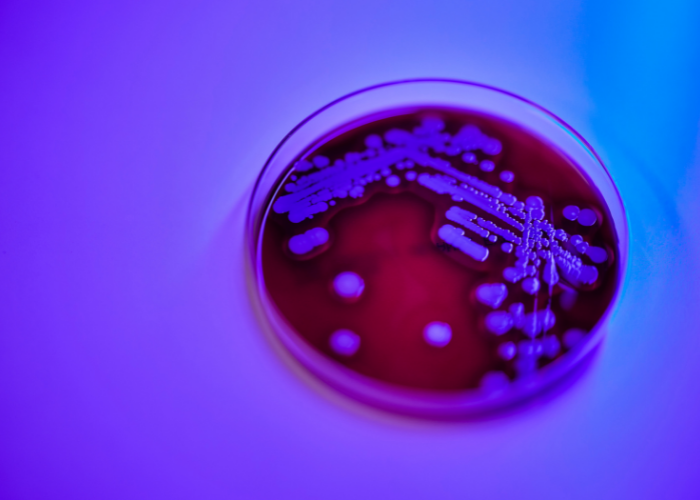
The discovery came about when, in the midst of their routine work, the scientists came across isolates that didn’t quite fit the profile of previously identified S. aureus pathogens.
Deciding to investigate further, they sent it for whole genome sequencing at the National Public Health Laboratory. Their suspicions were soon confirmed: the bacteria was a similar, but different strain.
The bacteria took two years to describe, the team said. Preliminary investigation suggests that S. singaporensis likely causes similar infections to other members of the complex, which are largely mild.
“But they do have the potential to cause a wide range of different infections,” Dr Chew explained. “So some [cases] can be a bit more superficial, with skin infections like dermatitis – but there’s also a risk of them invading and going deeper.”
Severe S. aureus infections can cause diseases such as cellulitis, osteomyelitis, endocarditis, or sepsis. These can be dangerous and even fatal.
The good news is that the new species appears to be less virulent that S. aureus, with fewer toxin genes. It is also susceptible to commonly-used antibiotics, unlike some S. aureus which have developed multi-drug resistance.
However, it is still in its early stages, Dr Chew added.
“Further investigations may highlight differences in terms of the propensity or ability of this species to cause infections, and whether there are any differences in their antibiotic resistance patterns,” he said.
Future strokes

The team emphasised that more research has to be done in order to gain a better understanding of the bacteria.
“With S. aureus, every one of us – at some time in our life – is sure to be infected. It’s impossible to escape,” said A/Prof Raymond Lin, Head and Senior Consultant, Microbiology, Department of Laboratory Medicine, NUH.
So the question that scientists must answer is whether S. singaporensis is likely to cause mild or severe disease – “whether it is more or less dangerous,” A/Prof Lin added.
For instance, the team’s assessment of the pathogen has so far been based on comparison with S. aureus. “But for all you know, S. singaporensis might carry its own set of peculiar virulence, we don’t know about that,” said Dr Jeanette Teo, Principal Scientific Officer, Microbiology, Department of Laboratory Medicine, NUH.
“So that could be research for the future. As time goes on, and we collect more and more of these isolates, we’ll better understand the biology of the disease.”
The team also stressed the importance of the whole genome sequencing technology – which the team used to identify the bacteria – in maintaining Singapore’s defenses against future disease outbreaks.
“With whole genome sequencing, we find that the species is novel and distinct, whereas with biochemical profiling, S. singaporensis is indistinguishable from other members of the complex,” explained Dr Teo.
This means that with biochemical profiling – the conventionally-used method for routine testing – the team would not have been able to identify the new pathogen.
Whole genome sequencing technology, which is present in most developed countries, was also used to identify COVID-19, the team added.
The team noted that having the tool available in Singapore laboratories means that scientists here will be able to identify the emergence of any new, harmful bacteria.
“The identification of this new species using the latest laboratory tools shows that our scientists in Singapore have the capability to investigate future emergence of new bacteria, which may cause outbreaks or severe disease,” said A/Prof Lin.
Dr Teo agreed, highlighting the importance of having access to such technologies – “without which, this new finding would not have been possible.”
In consultation with A/Prof Raymond Lin, Head and Senior Consultant, Microbiology, Department of Laboratory Medicine, NUH; Dr Chew Ka Lip, Consultant, Department of Laboratory Medicine, NUH; and Dr Jeanette Teo, Principal Scientific Officer, Microbiology, Department of Laboratory Medicine, NUH.